Publications & Case Studies
- All
- Publication
- Case Study
- Blog Post
- Poster
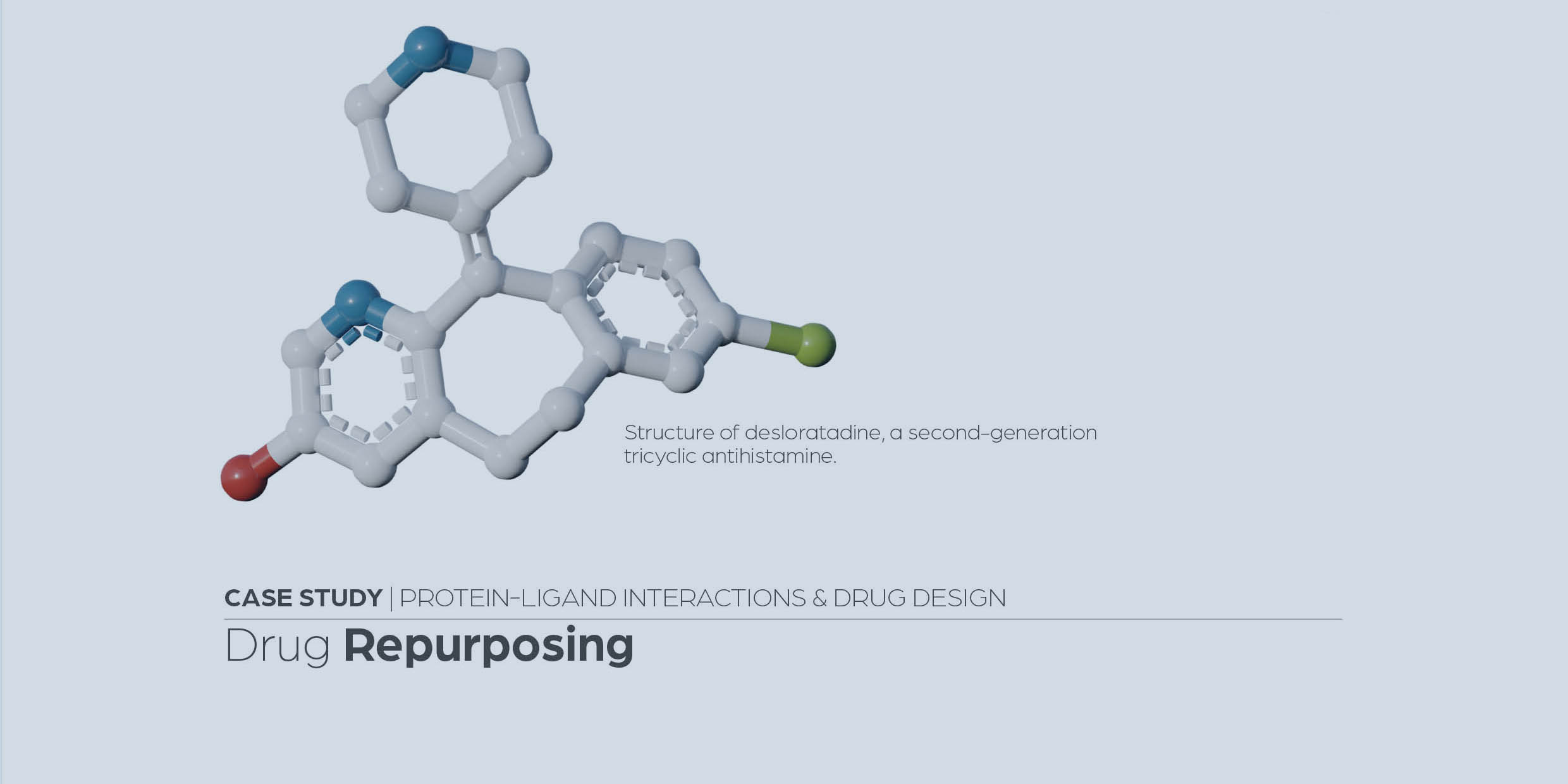
Identification and validation of fusidic acid and flufenamic acid as inhibitors of SARS-CoV-2 replication using DrugSolver CavitomiX

Hetmann, M., Langner, C., Durmaz, V. et al. Sci Rep 13, 11783 (2023).
https://www.nature.com/articles/s41598-021-83761-5
"Discovering dominant viral variants and their implications for host-protein binding."
Gruber C.C., Singh, A. & Resch V.
https://go.nature.com/3ZpObFN
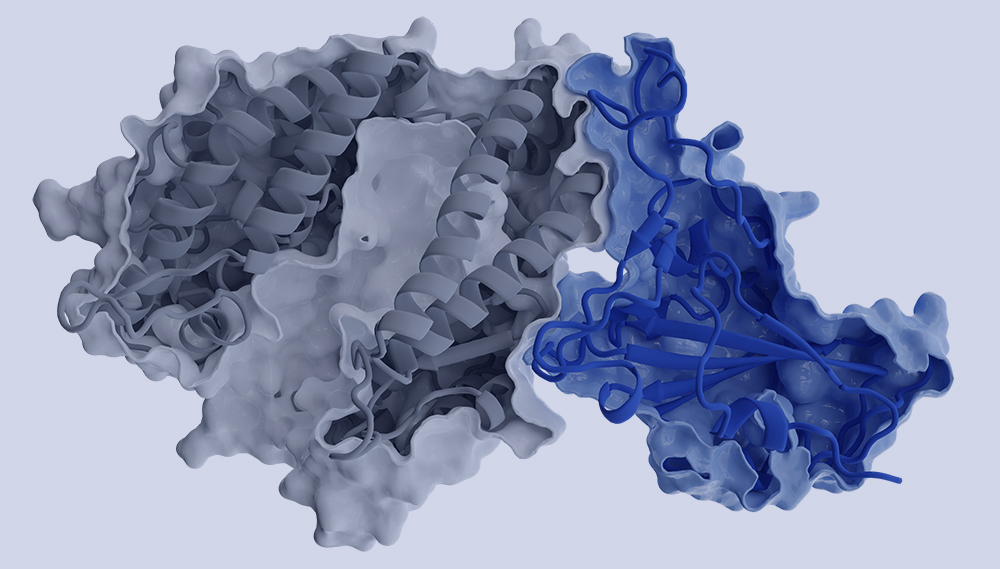
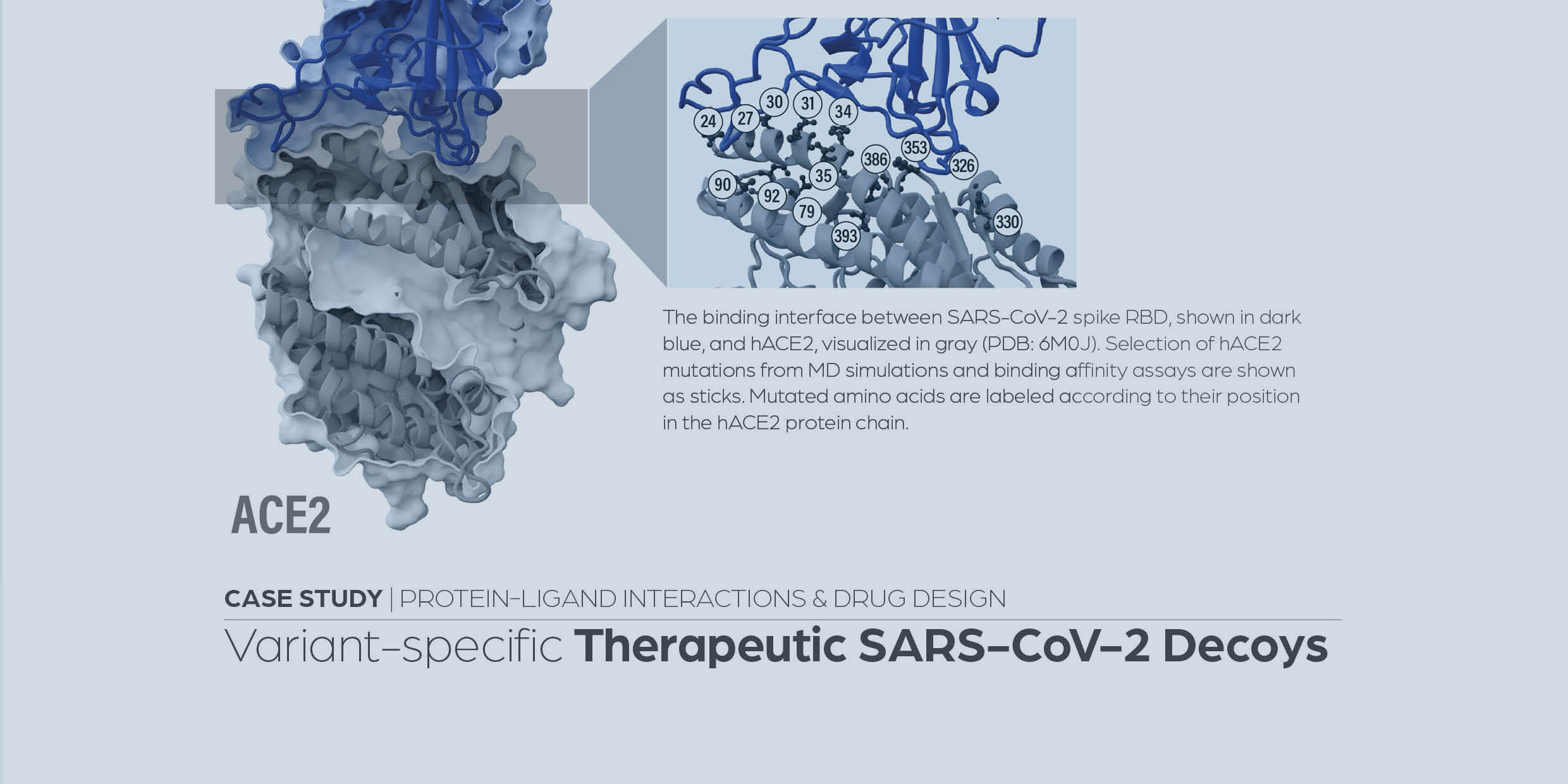
Optimizing variant-specific therapeutic SARS-CoV-2 decoys using deep-learning-guided molecular dynamics simulations

Köchl, K., Schopper, T., Durmaz, V. et al. Sci Rep 13, 774 (2023).
https://www.nature.com/articles/s41598-023-27636-x
Deceiving viruses: a blueprint for fighting future outbreaks and pandemics?
Gruber, C.C., Köchl, K., Krassnigg, A., Schopper, T., Resch, V.
https://go.nature.com/3QO7SUk
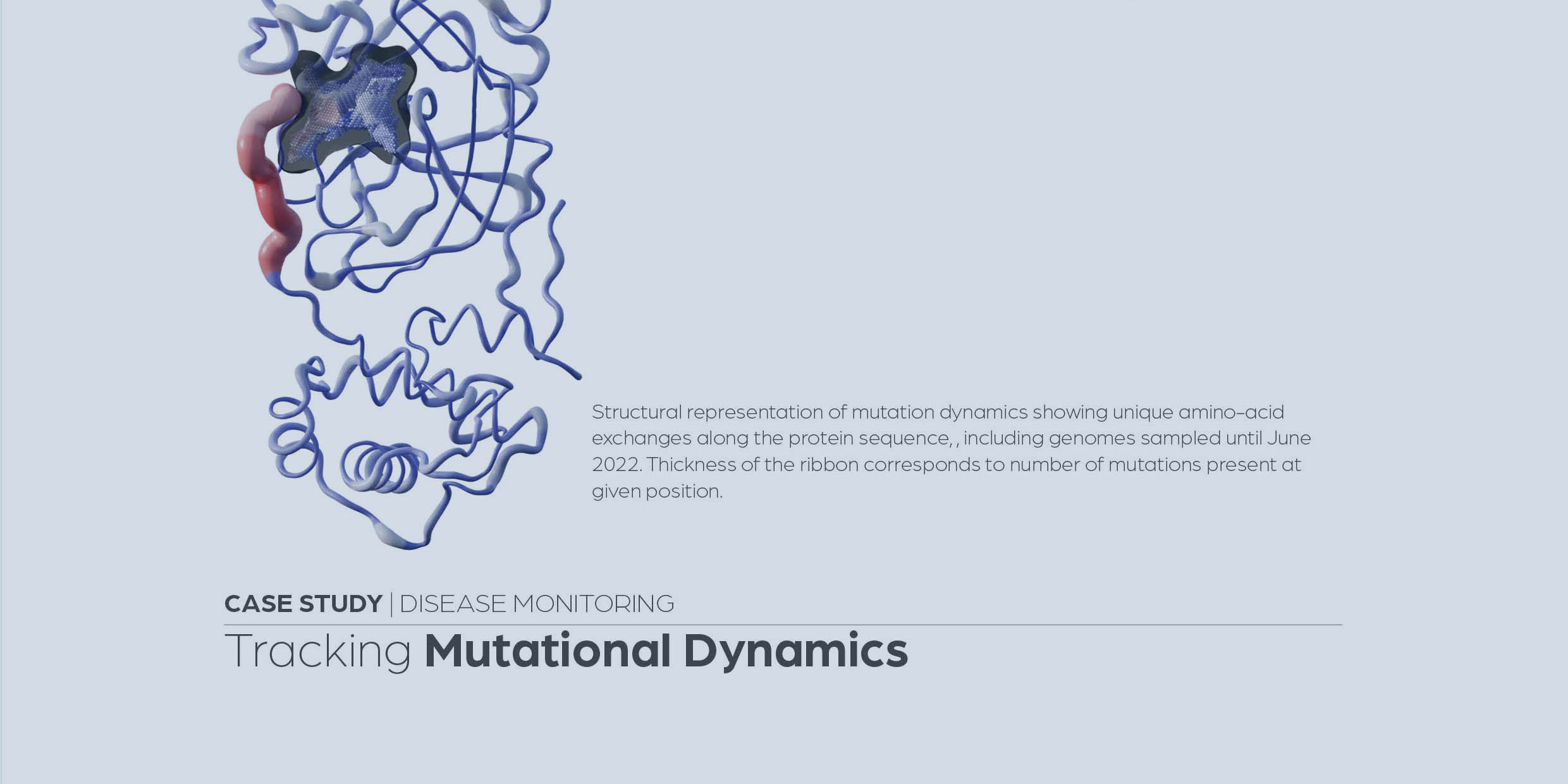
Recent changes in the mutational dynamics of the SARS-CoV-2 main protease substantiate the danger of emerging resistance to antiviral drugs

Parigger, L., Krassnigg, A., Schopper, T., Singh, A., Tappler, K., Köchl, K., Hetmann, M., Gruber, K., Steinkellner, G. and Gruber, C.C. (2022) Front. Med. 9:1061142.
https://www.frontiersin.org/articles/10.3389/fmed.2022.1061142/full
Preliminary structural proteome of the monkeypox virus causing a multi-country outbreak in May 2022

Parigger L, Krassnigg A, Grabuschnig S, et al. Research Square; 2022.
https://europepmc.org/article/ppr/ppr498427
Innophore's 3D point clouds provide a head start in monitoring emerging SARS-CoV-2 variant
Gruber, C.C., Resch, V.
https://go.nature.com/3NebrBk
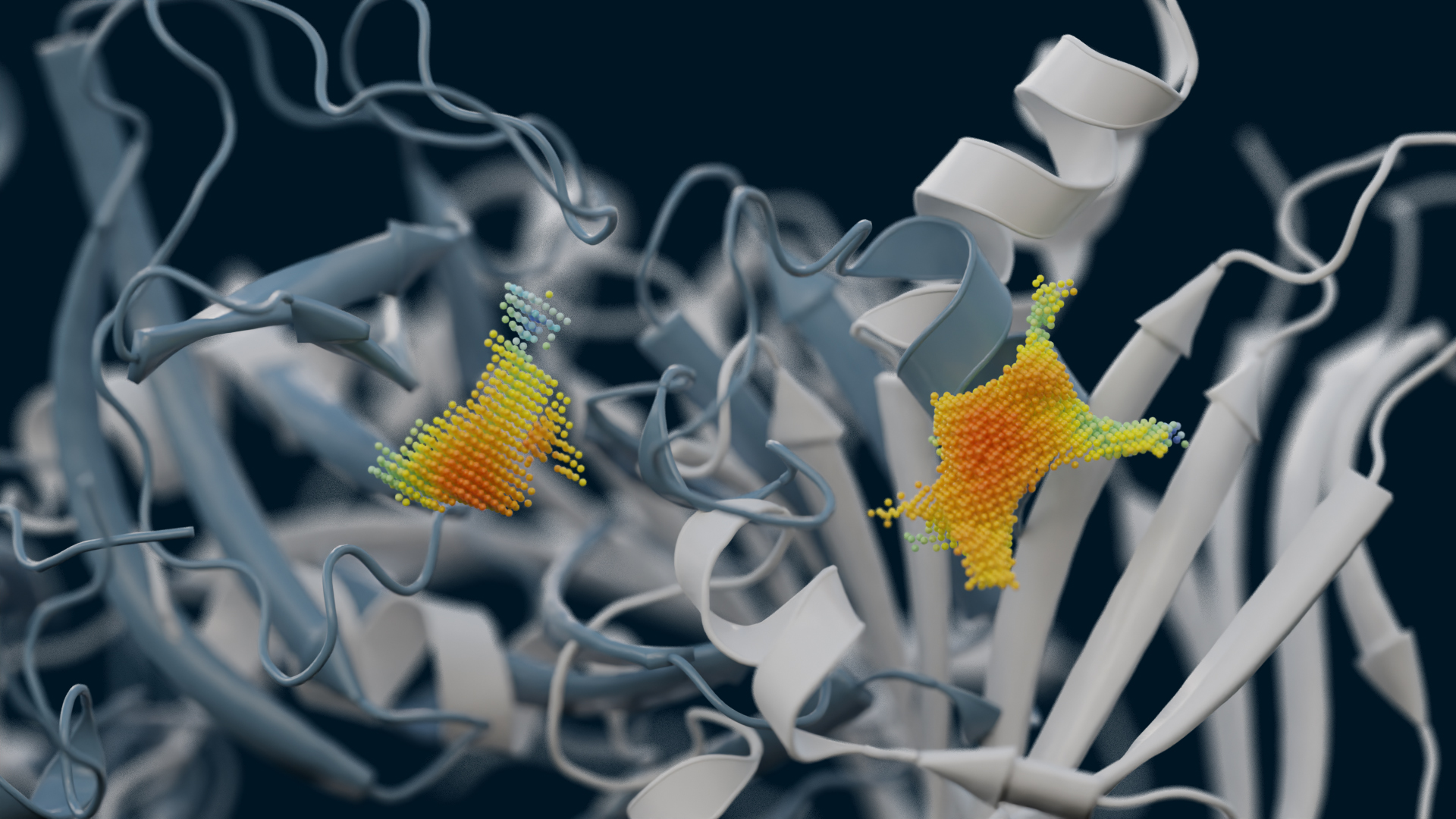
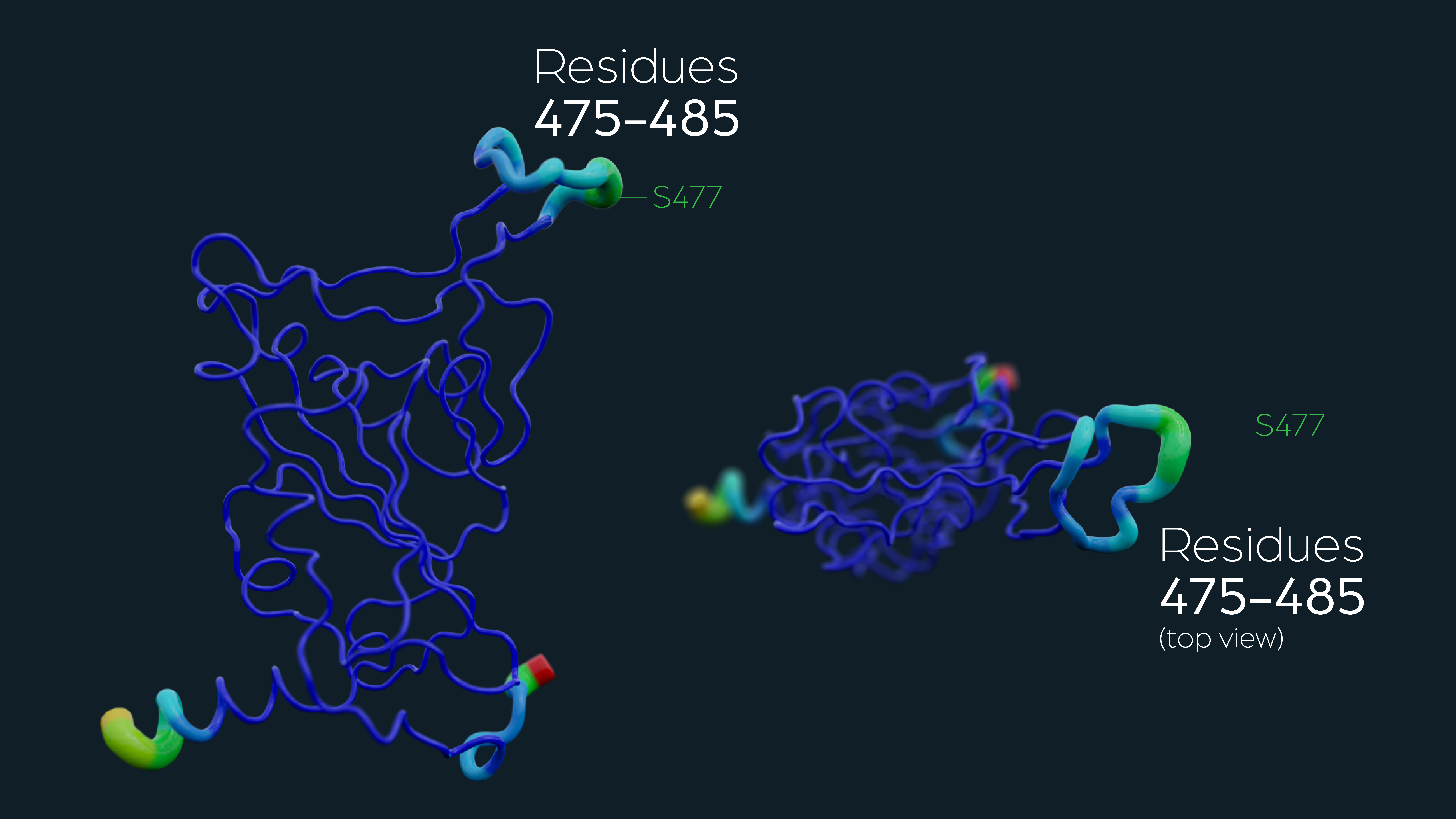
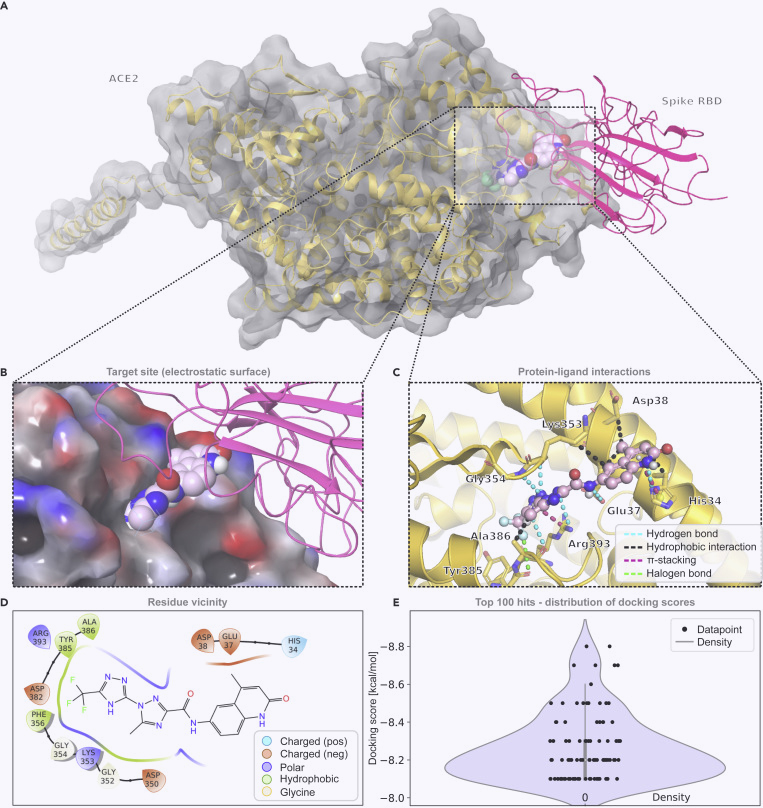
Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2

Singh, A., Steinkellner, G., Köchl, K. et al. Sci Rep 11, 4320 (2021).
https://www.nature.com/articles/s41598-021-83761-5
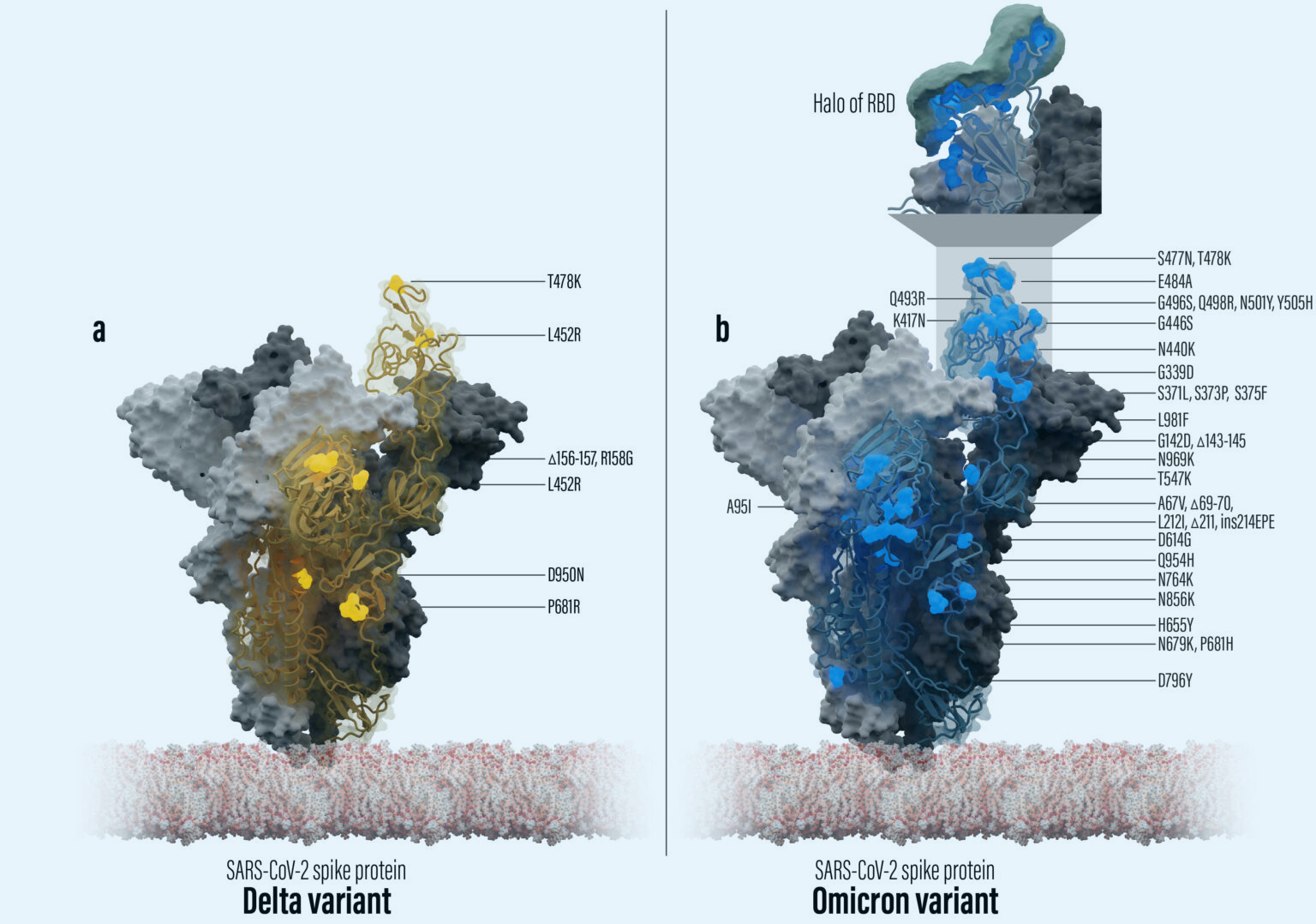
Structural bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild type reference

Durmaz, V., Köchl, K., Krassnigg, A. et al. Sci Rep 12, 14534 (2022).
https://www.nature.com/articles/s41598-022-18507-y
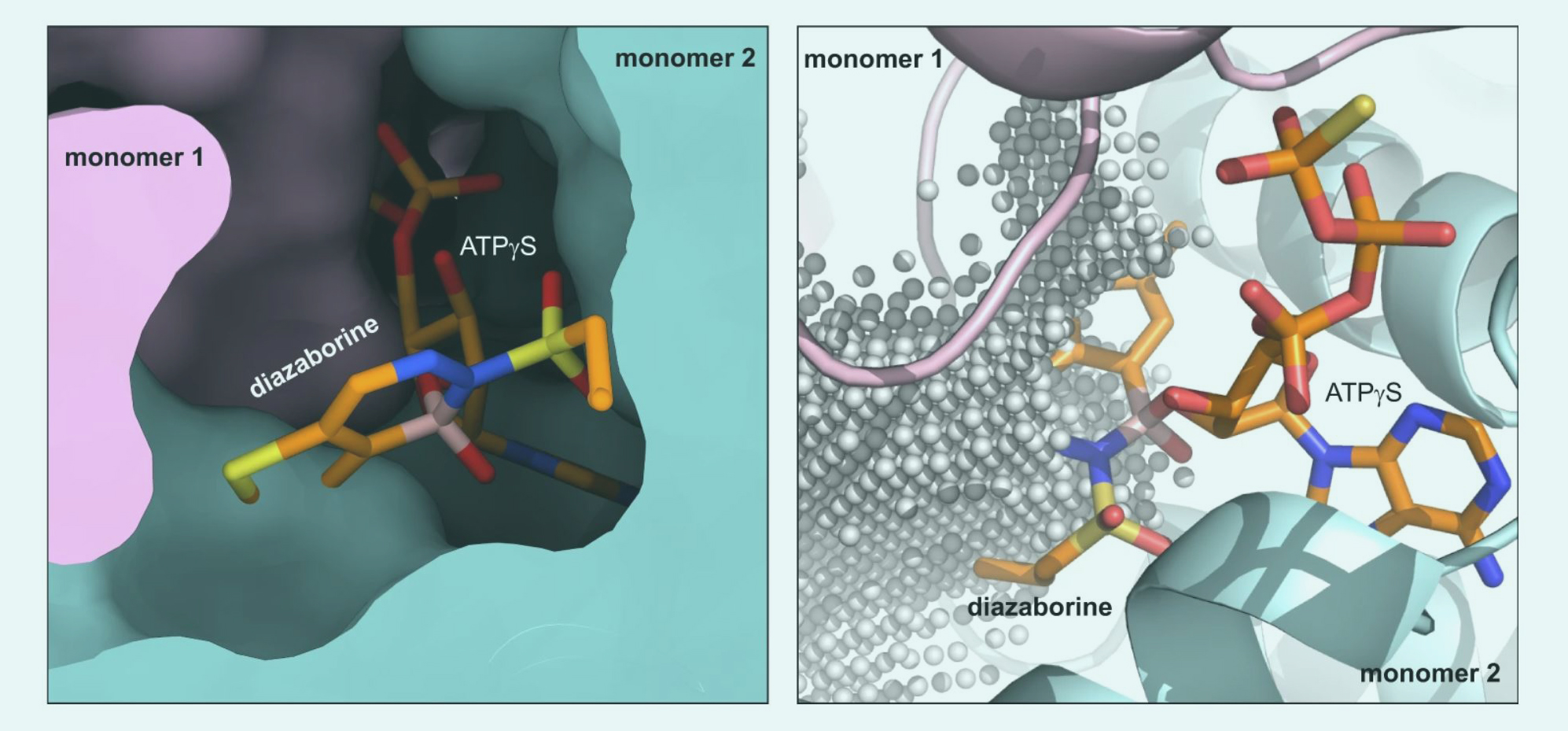
Structural basis for inhibition of the AAA-ATPase Drg1 by diazaborine

Prattes, M., Grishkovskaya, I., Hodirnau, VV. et al. Nat Commun 12, 3483 (2021).
https://www.nature.com/articles/s41467-021-23854-x
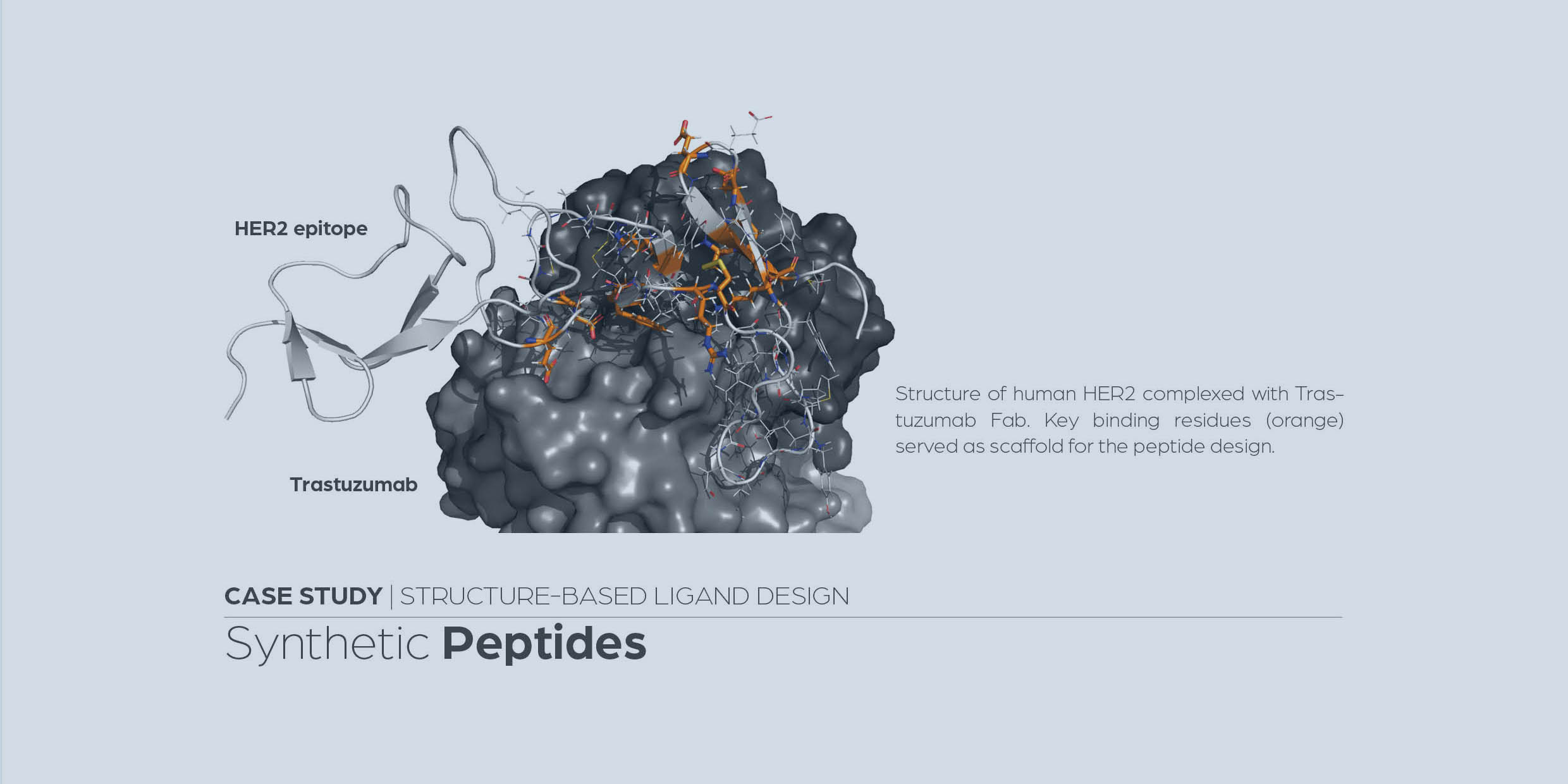
A multi-pronged approach targeting SARS-CoV-2 proteins using ultra-large virtual screening

Gorgulla, C., Padmanabha Das, K.M., Leigh, K. E. et al. iScience, Volume 24, Issue 2, 102021.
https://www.cell.com/iscience/fulltext/S2589-0042(20)31218-9
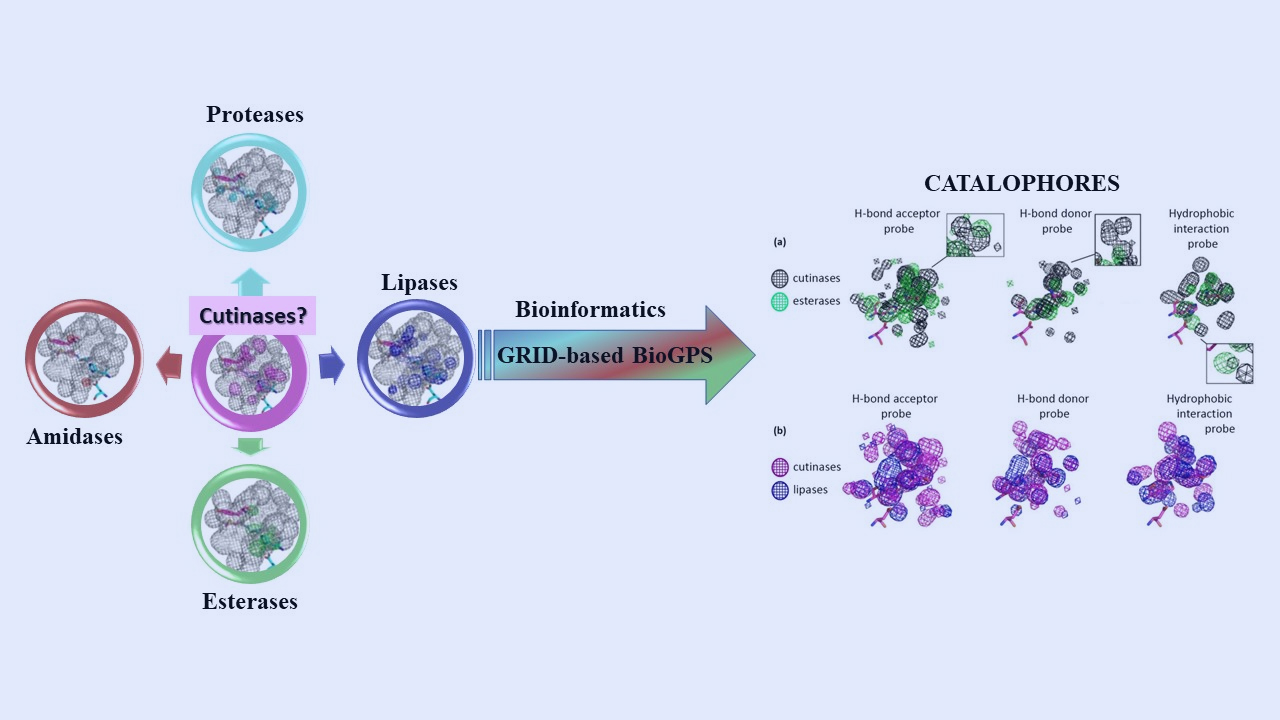
Criteria for Engineering Cutinases: Bioinformatics Analysis of Catalophores

Fortuna, S., Cespugli, M., Todea, A., et al. Catalysts 2021, 11, 784.
https://www.mdpi.com/2073-4344/11/7/784
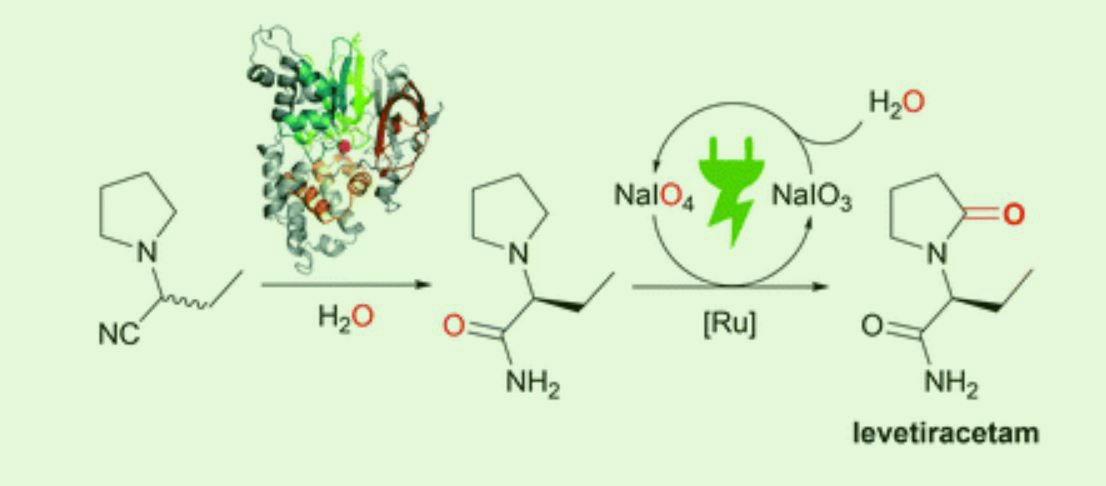
The sustainable synthesis of levetiracetam by an enzymatic dynamic kinetic resolution and an ex-cell anodic oxidation

Arndt, S., Grill, B., Schwab, H., Steinkellner, G. et al. Green Chem., 2021, 23, 388-395.
https://pubs.rsc.org/en/content/articlelanding/2021/gc/d0gc03358h
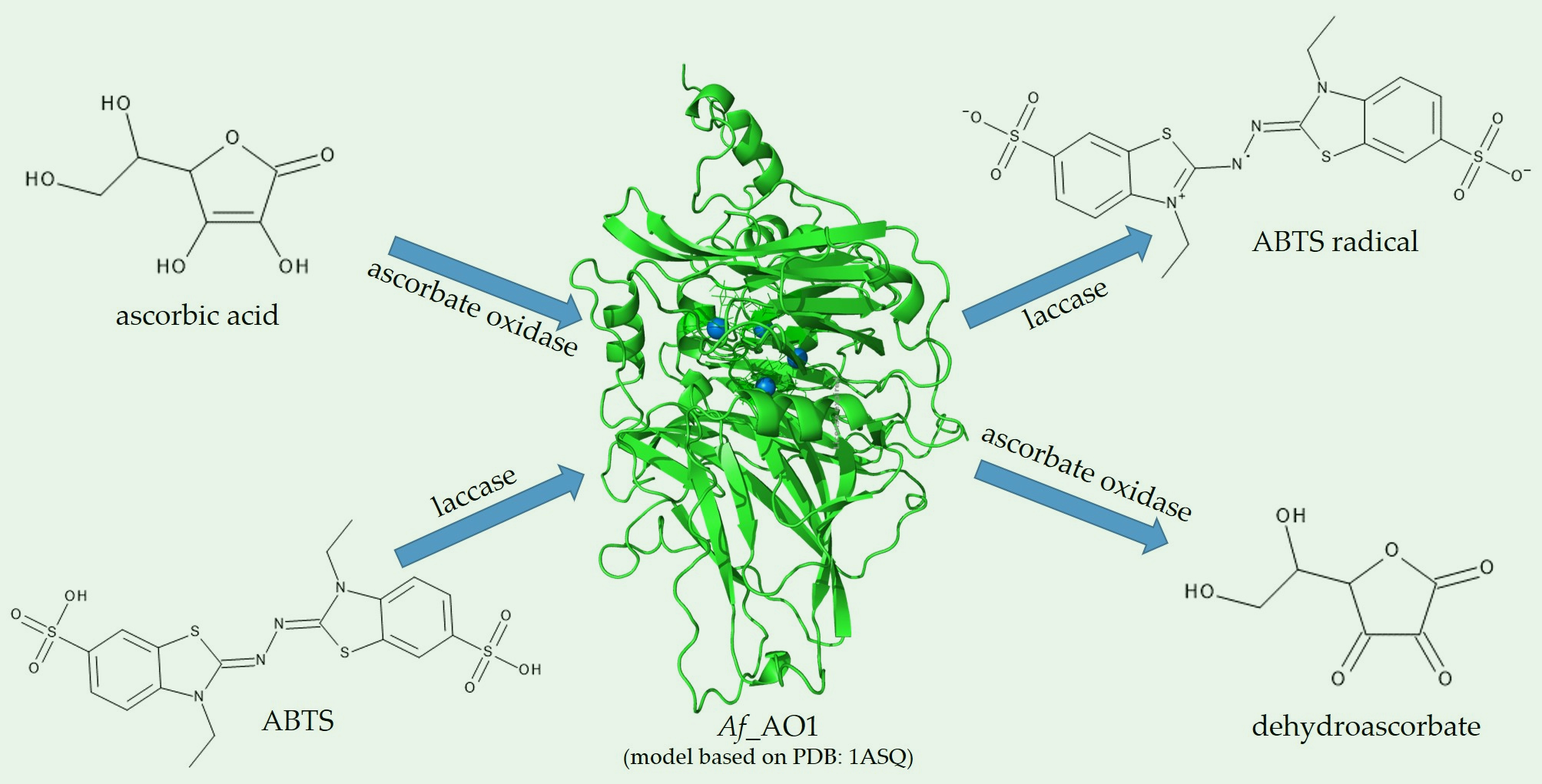
A Fungal Ascorbate Oxidase with Unexpected Laccase Activity

Braunschmid, V., Fuerst, S., Perz, V., et al. Int. J. Mol. Sci. 2020, 21, 5754.
https://www.mdpi.com/1422-0067/21/16/5754h
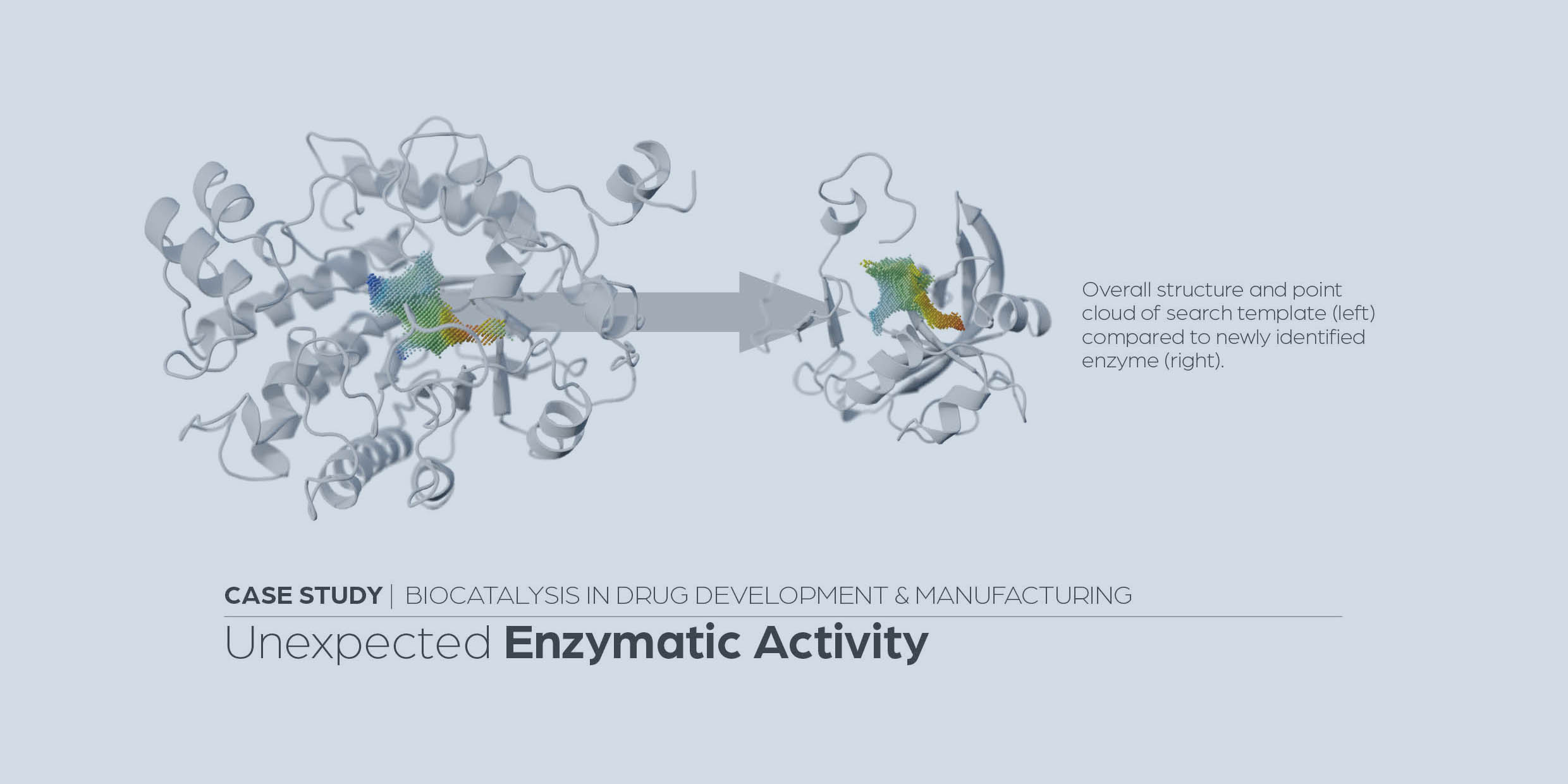
Identification of promiscuous ene-reductase activity by mining structural databases using active site constellations

Steinkellner, G., Gruber, C., Pavkov-Keller, T. et al. Nat Commun 5, 4150 (2014).
https://www.nature.com/articles/ncomms5150
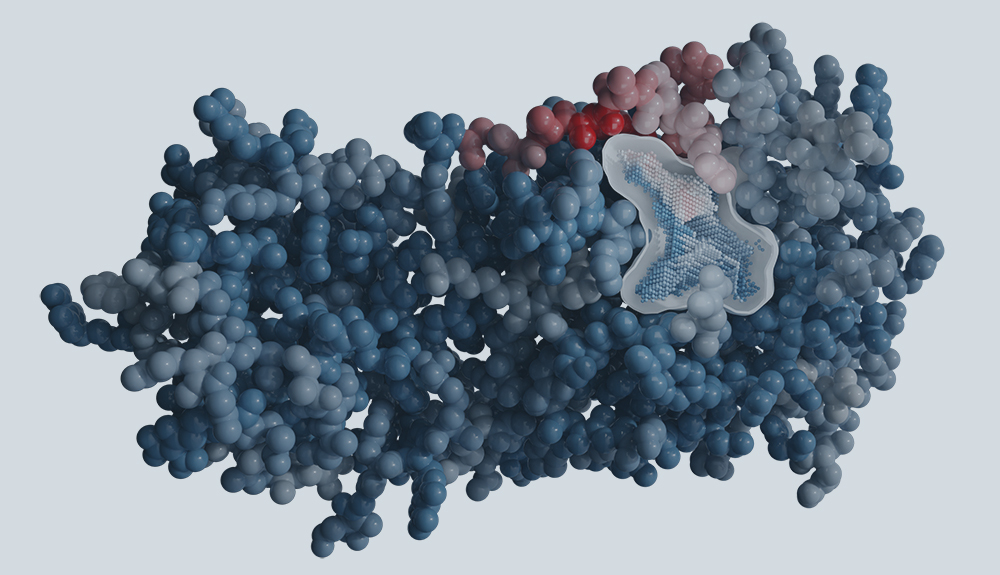
Determining novel enzymatic functionalities using three-dimensional point clouds representing physico chemical properties of protein cavities
Gruber, K., Steinkellner, G., Gruber C.C.
https://patents.google.com/patent/US20150302142A1/en
POSTER I virus.watch: Monitoring SARS-CoV-2 variants in cooperation with Amazon Web Services
Vedat Durmaz, Katharina Köchl, Andreas Krassnigg, Lena Parigger, Michael Hetmann, Amit Singh, Daniel Nutz, Alexander Korsunsky, Ursula Kahler, Centina König, Lee Chang, Marius Krebs, Riccardo Bassetto, Tea Pavkov-Keller, Verena Resch, Karl Gruber, Georg Steinkellner & Christian C. Gruber
POSTER I Variant-specific SARS-CoV-2 decoys: New perspectives for up-to-date COVID-19 treatment
Katharina Köchl, Tobias Schopper, Vedat Durmaz, Lena Parigger, Amit Singh, Andreas Krassnigg, Marco Cespugli, Wei Wu, Xiaoli Yang, Yanchong Zhang, Welson Wen-Shang Wang, Crystal Selluski, Tiehan Zhao, Xin Zhang, Caihong Bai, Leon Lin, Yuxiang Hu, Zhiwei Xie, Zaihui Zhang, Jun Yan, Kurt Zatloukal, Karl Gruber, Georg Steinkellner, Christian C. Gruber